Our approach
The science and technology behind our personalised insights
Science currently recognises 3–5 thousand gut-bacterial species, as technology, sequencing and awareness around the gut microbiome continues to advance, new ones are logged every month.
50,000+
We compare 50,000 profiled gut samples taken from two gold standard reference libraries, the Human Microbiome Project and the American Gut project and our own rapidly growing database.
30 - 40 trillion
The gut microbiome is home to roughly 30–40 trillion microbial cells. This figure is changing yearly as we discover more. Most of those microbial cells belong to only 3,000–5,000 gut-bacterial species.
150 - 200
The typical gut includes just 150–200 gut bacterial species drawn from that global pool of 3,000 - 5,000. Even closely related individuals share only a fraction of those species
We only work with accredited labs
ISO Accreditation 9001:2015 | 13485:2016
Our prefered method of testing
We analyse every sample with next-generation shotgun metagenomic sequencing because it unlocks the full story of your gut microbiome.
Instead of looking at a single genetic marker (like 16S rRNA), shotgun sequencing captures all DNA in the sample, telling us who’s there and what they can do.
That richer detail lets us pinpoint metabolic pathways, detect rare but important microbes, and link specific functions to health outcomes, giving you insights that are simply impossible with 16S alone.
Why is NGS shotgun metagenomic sequencing our preferred method of testing?
NGS Shotgun Metagenomic Sequencing gives the fastest, most complete snapshot of your gut.
Some sequencing systems such as NovaSeq are capable of reading 20–40 billion DNA fragments in just 13 hours, so it allows us to batch hundreds of samples overnight.
Shotgun sequencing captures all genes from all microbes including, bacteria, viruses, fungi, and archaea, unlike 16S tests that only scan one marker gene. - PMC
How accurate is it?
In mock community trials, shotgun sequencing pipelines correctly identify species with more than a 95 % precision, far exceeding 16S methods - Nature
Is shotgun metagenomics recommended for microbiome testing?
Yes, leading research and study guidelines now treat whole genome metagenomics as the reference method for comprehensive gut profiling - Nature
Major microbiome projects, including the two reference libraries we profile, the Human Microbiome Project and American Gut Project rely on the same approach, reinforcing its acceptance as the standard for microbiome testing.

Using large, high-quality reference libraries like the NIH Human Microbiome Project (HMP) and the American Gut Project (AGP) gives us a huge database of profiled samples.
These profiles give our machine-learning models two things, (1) Diversity, tens of thousands of well established genomes representing the full diversity of healthy and diseased guts worldwide. (2) Context, demographic, lifestyle and clinical metadata is captured that helps define what “normal” looks like across ages, diets and ethnicities and helps us to read between the lines.
What is a reference library?
A reference library is a large collection of data from previous, relevant studies that can be used for comparison and interpretation. Reference databases/libraries are essential in order to compare a patients results with published reference ranges to establish whether a result is within normal ranges and identify potential conditions.
Without a reference library, we would have nothing to compare your results to and essentially would be unable to paint a picture of what "normal" looks like.
How big are the reference libraries you use?
We use The Human Microbiome Project and the American Gut Project, both large microbiome studies that have helped to profile tens of thousands of gut microbiomes.
Our reference libraries and our own database includes around 50,000 profiled individual readings.
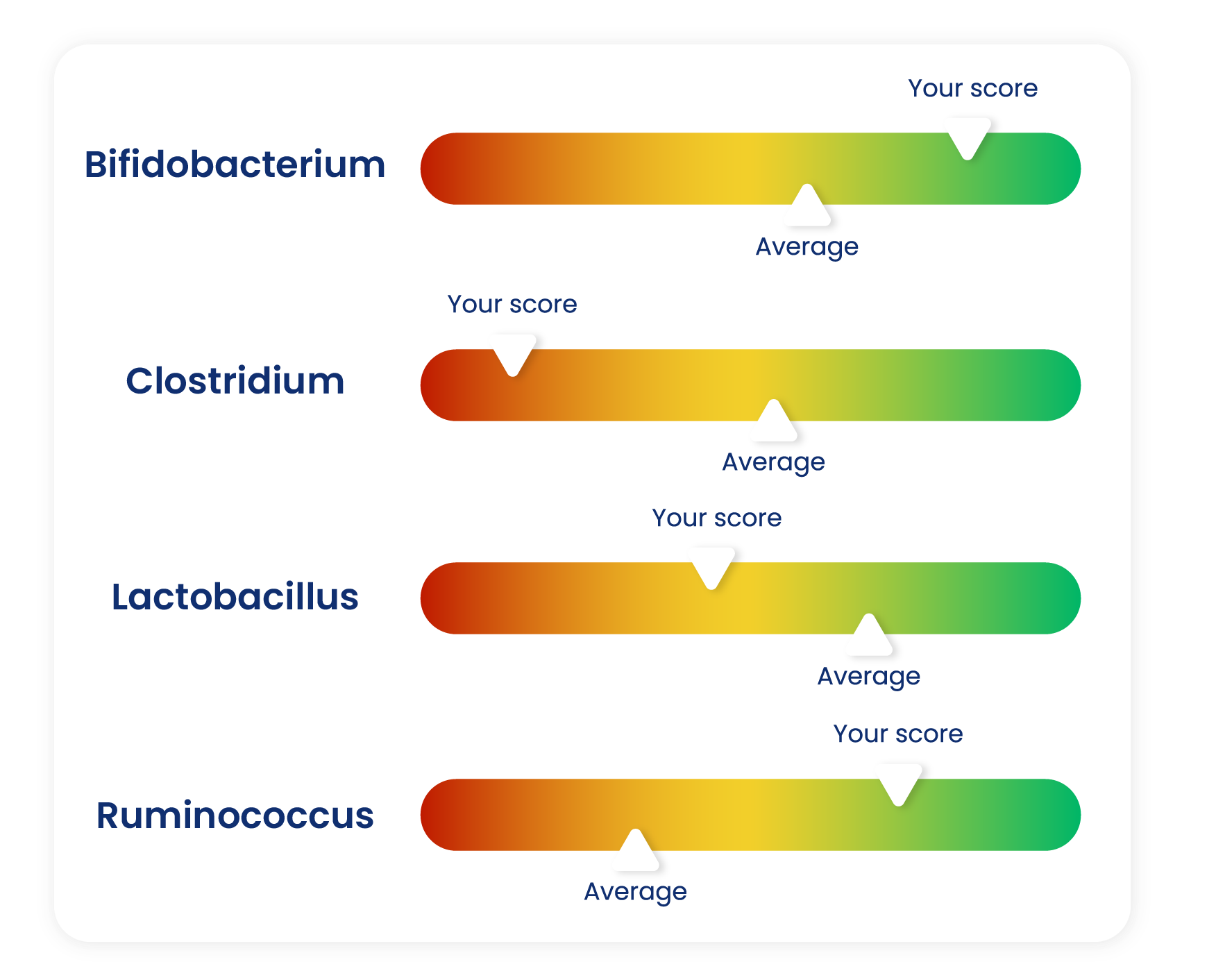
How do we compare readings with the data in the reference libraries?
We use machine learning models to spot combinations or patterns of microbes that are present in the reference cohorts, which enables us to profile your microbiome against that data.
After your DNA has been sequenced, it generates raw FASTQ files, which are digital records of every DNA fragment we have identified.
We upload these files to our secure machine-learning platform, which performs automated quality checks and profiling. Each microbe and gene is then matched against our two reference libraries, the Human Microbiome Project and the American Gut Project as well as our own profiled patient data so our models can see how your profile compares with tens of thousands of profiled samples.
Drawing on patterns learned from more than 50 000 profiled microbiomes and the latest nutrition studies, the system calculates condition likelihood scores and selects food or nutrient actions shown to shift the relevant bacteria.
After a final review from one of our medical experts, your personalised results and recommendations are published in a report, personal to you.
View the reportWhat is machine learning?
Machine learning (ML) is software that “learns” and looks for patterns from huge data sets, in our case thousands of gut profiles from projects like the Human Microbiome Project and the American Gut Project.
It spots relationships far too complex for manual statistics, letting us link specific microbe patterns to age, diet or health in seconds.
Advancements in technology such as Machine Learning has enabled further research in the gut microbiome.
How does Machine Learning interpret raw DNA into recommendations?
Machine learning compares your gut profile with tens of thousands of annotated samples in reference libraries such as the Human Microbiome Project and American Gut Project, then spots patterns linked to diet and health that scientists have already validated and are documented in those reference libraries.
For each flagged microbe or pathway, the algorithm pulls from peer-reviewed studies showing which foods or supplements reliably raise or lower it, for example, fibre types that boost butyrate producers, or polyphenols that curb pro-inflammatory species. - Pubmed MDPI
Do humans still review the results?
Yes, every report is reviewed by our team before registered dietitians and nutritionists check the personalised recommendations and nutritional suggestions before they are sent to you.
Our clinical studies
The results speak for themselves
Following a six-week personalised treatment plan, our patients report significant improvements in their symptoms and health.
Data taken from pilot study*